|
In October 2023, I had the wonderful opportunity to complete a two-month secondment at the CAMD (Computer Assisted Drug Design) lab in Urbino, Italy, under the supervision of Prof Giovanni Bottegoni. As a purely experimental biochemist, this was my first foray into an actual computational lab and I found the experience very enlightening. Of course, I had had previous introductions to computational methods and case studies from some of my Biochemistry courses, as well as from presentations at various conferences, and not the least from my fellow ESRs hailing from the computational side of the allostery field. However, in this case, I found the adage “experience is the best teacher” to be true. My hands-on use of tools such as Schrödinger and VMD showed me in a concrete way what it must be like to be a computational scientist, and I now have a better understanding of the typical workflow, as well as of the stakes of such work, including the balance between making faster versus more detailed simulations. Additionally, although my own training focused on proteins I was already familiar with from my experimental projects (the melanocortin receptors), several of my colleagues in Urbino were starting various new projects on completely different targets. This contrasts greatly with how experimental labs usually function with the focus remaining largely on related and/or functionally similar proteins across most projects. Despite this, at the CAMD lab, we could still help each other out, which fostered a real sense of camaraderie and a greater understanding of each other’s goals. I believe, now more than ever, that the future of science, and drug discovery in particular, truly relies on, not just one or the other, but the symbiosis of both computational and experimental methods. Thus, it is crucial to understand the possibilities as well as the limits of both, in order to lead to optimal collaborations.
Finally, I would like to highlight the backdrop of my journey into the computational world: the gorgeous city of Urbino, relatively unknown internationally, but a marvel of Renaissance arts and architecture, as anyone attending the recent ALLODD conference there surely noticed. I was happy to return once more and have the chance to say a quick hello to my former colleagues at the CAMD lab.
0 Comments
More than two years have passed since I joined ALLODD and I am now delving into scientific conferences with one objective: share. Conferences are great opportunities to meet with people from your field and I was really amazed by the kindness of people who are always happy to discuss scientific ideas but also share their thoughts on career path. Among numerous insights, here are the most significant tips:
Be curious As scientists, we enjoy uncovering new problems and devising innovative solutions. Cultivating a curious mindset involves continuous learning and staying connected with the latest advancements in your field and beyond. Embracing curiosity fuels your drive to seek out new challenges and opportunities with happiness. Be focused Establish clear career goals and remain resolute in your pursuit of them. A focused approach ensures that you stay on track and work diligently towards achieving your ambitions. By staying unwaveringly focused, you can navigate through the complexities of your scientific journey with purpose and determination. Build a network Whether your professional aspirations lead you towards academic pursuits or the industry, the value of building a strong network cannot be overstated. By fostering a network, you can create a supportive ecosystem that is vital for support, guidance, and opportunities. Funnily, I met at these conferences people who worked in Darmstadt, Germany (where I am currently living) or even in Clermont-Ferrand, France (where I studied). In France, we have a saying “Le monde est petit” (the world is small). Be unique Last but not least. Each scientific journey is inherently distinct, shaped by individual experiences and perspectives. Embracing your unique attributes and experiences is a powerful way to differentiate yourself and pique the interest of others. By showcasing your uniqueness, you can carve out a compelling narrative that sets you apart in the scientific community On 27th February- 1st March 2024 we ESRs had the great chance to get trained through the 3rd ALLODD Training School in the J&J campus in Beerse, Belgium “IPR Training for Researchers & ESR Presentations on the Progress of their Research”.
It was a great opportunity to get in touch with the industrial world once again and experience also the personal stories of some of the speakers alongside training and information. It has been very interesting to get to know how the “big pharma” is trying to support innovation through several partnerships and founding even physical ones like the JLABS and get in line with the new guidelines like the project for the reduction of emission which Johnson will try to reach in 2026. Having a geothermal power plant within a campus is surely quite impressive! Also, the attention to animal health care, it’s something I am sure people are expecting to see more and more applied and this is also a great achievement. Because if it is true that (unfortunately I would say!) they are still required, it is also true that our efforts can be applied even more in this sense, after all (trying to relieve pain isn’t what we should be trying to focus on?) I just hope we will see more and more of this in future and worldwide! Alongside this, we had the opportunity to get in touch with very interesting talks like intellectual property and Project management and partnerships on the first day and scientific ones on Car-T cells therapy, PROTACs and the kinases. In sum a well-compelled series of talks ranging from biology to chemistry, medicinal chemistry and more! But this was not all, on the third day we had to practice (and I would dare to say re-learn!) how to introduce ourselves rapidly but exhaustively in a 4 and then in 1-minute pitches trying to catch as much as possible, which is surely not an easy task! Yet it is interesting to get a little detached from our daily way of expressing ourselves within the scientific community and get more to the general public. After all, as George Orwell has greatly shown in 1984 behind simplification there can be a huge work, but I would add sometimes this can also be very beneficial! All this simplification process is in fact one of the basic sets for effective communication in which a key role is also relying on intonations, gestures, and looks and is always very important to keep in mind! We cannot be thankful enough to ALLODD and Janssen Pharmaceutical for this experience and further opportunity and as learned from hearing the story of Janssen himself, proceed even with more enthusiasm cause the clock is ticking and “patients are waiting”. And after all, we are scientists, and we hope to give our contribution also to help! Lessons in Chemistry is a miniseries that tells the life story of chemist Elizabeth Zott in the 1960s. Unfortunately, she is not a real person, and chemistry only serves as a secondary character, blending with the atrezzo. However, I’m still going to take advantage of this blogpost to quickly talk about my real passion: TV shows.
In this case, this show is based on a fictional book of the same name, so the leading scientist didn’t exist. Nevertheless, the situations she goes through and the life she ends up living can easily be assumed to be an accurate representation of those times. Telling the story of a female scientist in 1960s US, it obviously deals with misogyny and the role it played in hindering and even truncating many women’s careers, still reverberating to this day. And to this viewer-reviewer, the interesting part is that these topics are depicted in a markedly refreshing way. Although none of it is novel, instead of softening, dumbing down, or sugarcoating situations of rampant sexism, for example, I found that the scenes are laid out with such rawness that they felt truly authentic. In an age of mass-produced entertainment extruded by Netflix and the algorithmic feeling of everything it puts out, in Lessons in Chemistry I could appreciate a specific and differential creative vision and drive; a human heart behind it, in short. I personally like to imagine that such a human is Brie Larson herself, who serves as the lead actress and as a producer, and whom I found to have delivered an amazing performance, supported by a very effective cast. Sprinkled on top, we get some corny lines supposedly pronounced during the inception of nucleic acid research, and others about the chemical reactions that take place when food is being cooked. Both are blurted out by the actors as meaningless strings of words that they were told to memorize, but I didn’t pay attention to their scientific rigour and also it doesn’t matter. Chemistry here is a background character used to arrive to the conclusion that we can’t control for every variable, but uncertainty may lead us to places we’re happy in. Ew. Also as a background element used to move the story forward, our heroine gets entangled (ew) with a man who is a fellow scientist. But this character diligently reinforces the accuracy of the “men written by women” online phenomena, as there’s no way this person ever existed or even could exists nowadays, but that may be a topic for another blogpost. Overall, Lessons in Chemistry amounts to a non-scientific well-made and enjoyable show, and its complete first and only season just wrapped up and is available on Apple TV+. Developing Time Management Skills: A Path to Greater Productivity - a blogpost by ESR10 Sonja Peter16/10/2023 In the fast-paced world we live in, one secret to success is to manage time efficiently. During our ALLODD meeting in Budapest [1], we had training on how to increase productivity through efficient time management. In this blog post, I would like to share 7 key points: 1. The Three Pillars – environment, mind, schedule: Organise your working space to minimize distraction, clear your mind from thoughts by taking notes, and structure your day using the 18-minute/day method. The 18 minutes consist of 5 minutes dedicated to planning the day, 8 minutes to refocus every working hour, and 5 minutes at the end of the day for reflection. 2. Estimate time: To estimate work durations, use historical data or tools like the GANTT chart (Figure 1 A) to analyse the critical path and to spot bottlenecks that require the most time. 3. Feedback: Find the correct timing to ask for feedback to ensure that the work is following the correct focus required for completion. 4. Prioritization: A distinction between important and urgent jobs helps in setting priorities (Figure 1 B). Important and urgent tasks such as responding to accidents, giving a short minute presentation for important collaboration opportunities, or responding to a review are called firefighting tasks. These tasks are generally managed well. More challenging is to balance reactive and proactive tasks. Reactive tasks such as responding to e-mails or spending time on social media are loud, meaning we are notified regularly, while long-term proactive tasks such as career development or working on a paper/project are quieter. It is important that moderately urgent tasks do not get overshadowed by urgent ones. 5. The Power of Habit: Understand that habits play a significant role in time management. Cultivate good habits that align with your goals as this can help to master periods of large change such as moving to another country or working for some time at another organization/university. 6. Small Changes: Don't underestimate the impact of small changes in your routine. Consistency is key. 7. Elephant vs. Rider: Imagine your conscious mind as the rider on top of the elephant (habit) (Figure 1 C). It needs time and conscious efforts to guide your habits toward effective time management. Many of the points were known to me before the training, but the refresher reminded me to incorporate these techniques into my daily routine. I started to integrate the refocusing every hour to help me improve my productivity in the last two weeks. [1] Time management, Trainer: János Balázs KISS, Mindbeat, September 2023 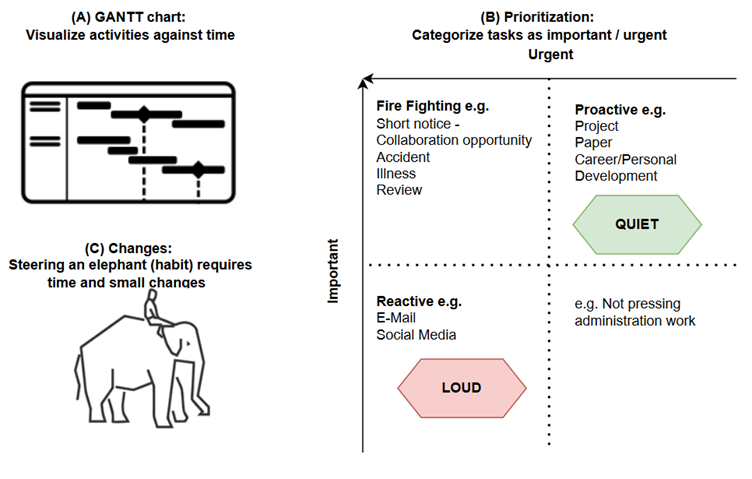 Figure 1. Overview of tools for time management. (A) GANTT chart can be used to plot the tasks against time and identify critical steps that require time. (B) Tasks can be categorized into importance and urgency. (C) The consciousness is the rider, while the elephant is the habit. The rider needs to make small changes to the moving elephant. Developing Time Management Skills: A Path to Greater Productivity. I've reached the exact midpoint of my fellowship, and it has been an incredibly transformative 18 months! From relocating to three new countries, connecting with ALLODD members, and going deeper into my project, it has been a whirlwind of growth and exploration. The experiences I've encountered during this time have undoubtedly broadened my horizons, and there's still so much ahead to embrace!
Two topics that have particularly caught my attention recently are machine learning and AI in science. During my two-month secondment in Athens and the insightful conversations with my peer there (thanks again for our discussions 😊), I've become a bit less naive about the topic. While I've used chatGPT for travel tips before, I now recognize that machine learning and AI have a profound impact on the future of science. As more and more pharmaceutical companies are venturing into AI and machine learning to transform their processes, embracing these new technologies is crucial also for bench scientists like me. While the predictions I've received might not always be accurate, the real power lies in the way these tools streamline and narrow down possibilities. In the fast-paced world of science, time is a precious asset, and AI serves as a valuable ally, empowering scientists like me to make more informed decisions, faster. Looking ahead to the next steps of my PhD journey, I'm filled with excitement and curiosity. The best is yet to come! Chemical space is vast. So vast, that screening even a relatively small portion of this space within a drug discovery campaign is close to impossible. Just considering molecules with 30 heavy atoms consisting of carbon, oxygen, nitrogen, and sulfur we would find more than 1060 possible combinations [1] - a number that make even industry-scale million-member libraries of compounds appear small. While these libraries are still successfully used in high-throughput screens, a different approach, coined fragment-based drug discovery (FBDD) over 20 years ago [2], makes use of collections of so-called fragments with lower molecular weight (<300 Da) and a limited number of structural features [3]. The small size of the fragments drastically decreases the number of possible molecules and enables FBDD approaches to cover broader chemical space more efficiently. Due to their low complexity and low number of ‘non-essential’ atoms, screening fragments usually yields higher hit rates of molecules whose interactions with the target are defined by only one or two high quality contacts and high ligand efficiency (binding free energy per heavy atom). Accordingly, FBDD routines can be instrumental in finding druggable ‘hot spots’ and defining starting points for lead discovery. Yet, with small size often comes low affinity. Probing weak protein-fragment interactions demands for highly sensitive biophysical techniques, such as NMR, X-ray crystallography, surface plasmon resonance (SPR) or thermal shift assay (TSA), which often depend on availability of the purified target and yield only limited information on the effect the targeted site has on the functionality of the protein in a physiological environment [3]. The latter is particularly important for allosteric proteins considering the diverse mode of actions allosteric modulators can exert. Functional cell-based assays would allow for such a read-out in the absence of soluble target protein but are limited by the high concentrations of the fragments necessary to induce a measurable effect. One way to mitigate the challenges of detecting low affinity fragments, is so-called covalent tethering. In equipping fragments with a warhead that can form a covalent bond close to the target’s binding site, tethering increases the effective local concentration and therefore also apparent affinity. Reversible disulfide-bond formation between a thiol-modified fragment and a free naturally occurring or genetically inserted cysteine on the target is probably the most frequently used method to trap interacting fragments at the target site [4]. Most prominent example for this approach is the discovery of a covalent allosteric inhibitor for K-Ras G12C [5]. Recently, this approach has also enabled validation of the mode of action of a low-affinity allosteric fragment for PTP1B [6]. However, cysteines often represent critical sites for disulfide bond formation, posttranslational modifications and can be highly abundant throughout the target and off-target structures. Accordingly, disulfide-based tethering can often not be applied in cell-based assays and requires careful optimization and validation. In particular, if the cysteine is introduced by mutagenesis. In a recent paper, Mattheisen et al. [7] present an alternative elegant approach that leverages bioorthorgonal coupling chemistry to tether ligands to the class A GPCR C-C chemokine receptor 5 (CCR5) for functional fragment screening in live cells. Due to its critical role in HIV-1 pathogenesis, CCR5 has been rigorously characterized in terms of structure, function and druggability. Likewise, allosteric antagonists and the corresponding allosteric binding site have already been identified making it an ideal system to establish the proposed method [8]. While no large-scale fragment screening is conducted, the study provides a proof-of-concept and explores important considerations when establishing such a system. 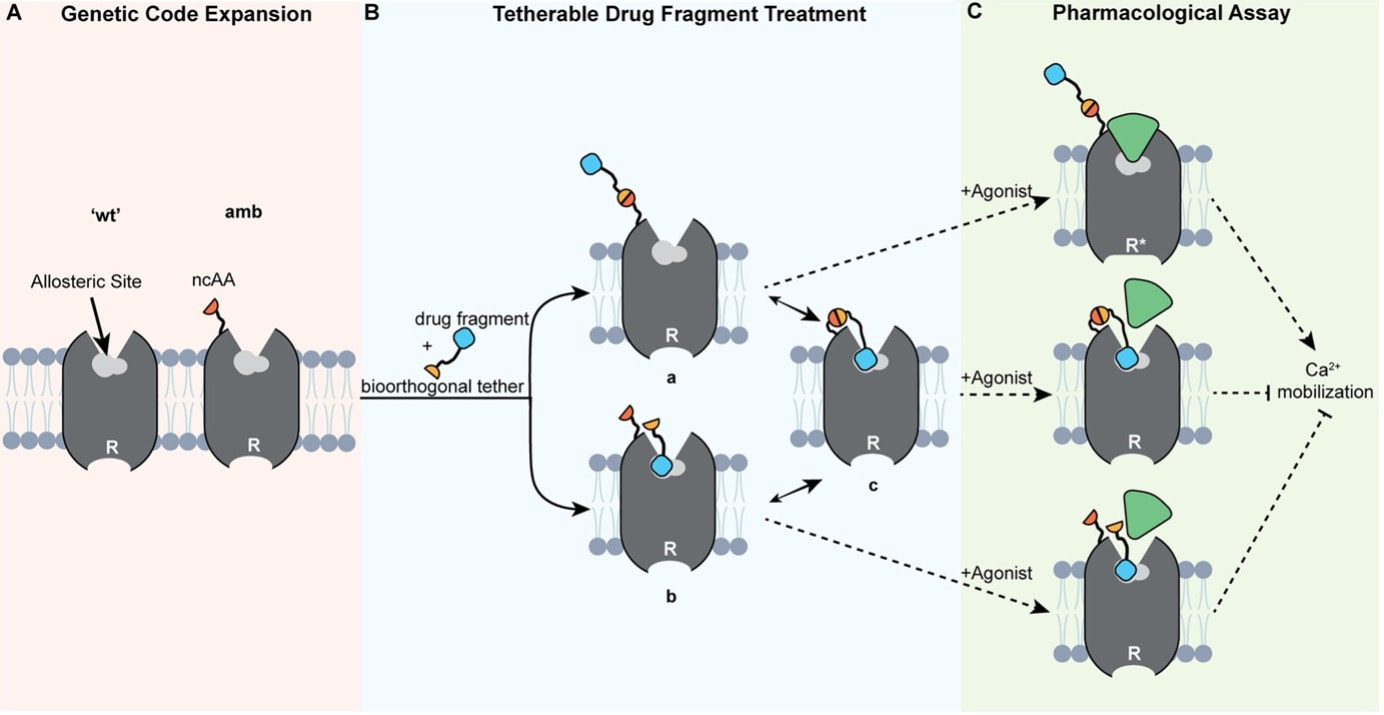 Figure 1: Setup to test antagonist activity of heterobifunctional ligands upon bioorthorgonal covalent tethering to a reactive handle incorporated into the GPCR CCR5. A) The reactive handle (here a non-canonical amino acid, ncAA) is incorporated into the target receptor close to the allosteric pocket by genetic code expansion. B) The heterobifunctional ligand is added and either a) first reacts with the handle, followed by binding or b) first binds to the pocket and then reacts or c) both. C) Potency is the assessed by adding an agonist and measuring activity of the receptor (here by Ca2+ mobilization). Figure and caption were adapted from Mattheisen et al. [7]. The system consists of a chemical handle that is incorporated into the receptor close to the allosteric binding site (Figure 1A) that can react with a reactive functional group under bioorthorgonal conditions compatible with mammalian cell culture (as introduced by the Bertozzi lab [9]). The reactive warhead is linked to a ligand via a PEG spacer, allowing a potential, now covalently coupled, antagonist to reach into the allosteric pocket which lays deeply buried in between the receptor’s transmembrane helices (Figure 1B). Functional response upon reaction with this heterobifunctional ligand is then probed by assessing cellular response to an agonist (Figure 1C). Using a combination of molecular dynamics simulations and subsequent experimental validation, Mattheisen et al. optimize assay conditions and controls by varying insertion points of the chemical handle in the receptor, warhead chemistry and linker length. Throughout the optimization process, the FDA-approved CCR5 antagonist maraviroc (mvc) is used as an active antagonist on the heterobifunctional ligand, which worked nicely for showing the theoretical potency increase the covalent tethering yields. So far so good. But mvc is a highly potent antagonist with nanomolar activity after all. So how would this system perform for lower affinity compounds, such as fragments? To test the capability of the system to distinguish between non-binders and low-affinity binders, six heterobifunctional ligands carrying low affinity mvc analogues were synthesized and tested in two cell-based assays, one testing for ability to block agonist activity and the other for blocking labeling of the reactive handle by a warhead-fluorophore conjugate. Intriguingly, this strategy revealed that only those heterobifunctional ligands whose low affinity mvc analogues show antagonist activity would also block labeling of the reactive handle by the fluorophore. Moreover, only the receptor containing the reactive label was inhibited. This is an important observation, since it indicated that on the one hand, the potency of low affinity compounds is effectively increased by covalent tethering and that on the other hand, affinity of the fragment for the pocket is the driving force for the reaction. In summary, the paper presents an innovative approach for live-cell functional screening of low-affinity compounds that adds another valuable method to the drug discovery toolbox for pharmaceutically interesting GPCR targets. However, some open questions remain: How easily is this system transferable to other receptor classes? How does the system perform in large-scale fragment screenings as shown for disulfide-tethering? How does it perform in a direct comparison with biophysical fragment screening in terms of hit-rates and false-positive rates? Can it facilitate fragment-to-lead discovery? References
1. Bohacek, R. S., McMartin, C. & Guida, W. C. The art and practice of structure‐based drug design: A molecular modeling perspective. Med. Res. Rev. 16, 3–50 (1996). 2. Shuker, S. B., Hajduk, P. J., Meadows, R. P. & Fesik, S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science 274, 1531–1534 (1996). 3. Erlanson, D. A., Fesik, S. W., Hubbard, R. E., Jahnke, W. & Jhoti, H. Twenty years on: the impact of fragments on drug discovery. Nat Rev Drug Discov 15, 605–619 (2016). 4. Erlanson, D. A., Wells, J. A. & Braisted, A. C. TETHERING: Fragment-Based Drug Discovery. Annu Rev Bioph Biom 33, 199–223 (2004). 5. Ostrem, J. M., Peters, U., Sos, M. L., Wells, J. A. & Shokat, K. M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). 6. Keedy, D. A. et al. An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering. Elife 7, e36307 (2018). 7. Mattheisen, J. M. et al. Bioorthogonal Tethering Enhances Drug Fragment Affinity for G Protein-Coupled Receptors in Live Cells. J Am Chem Soc (2023) doi:10.1021/jacs.3c00972. 8. Tan, Q. et al. Structure of the CCR5 Chemokine Receptor–HIV Entry Inhibitor Maraviroc Complex. Science 341, 1387–1390 (2013). 9. Prescher, J. A., Dube, D. H. & Bertozzi, C. R. Chemical remodelling of cell surfaces in living animals. Nature 430, 873–877 (2004). Just last week, I attended my very first conference: an ALLODD one, no less! Held in the understatedly beautiful city of Strasbourg, this event was my first chance to finally meet my fellow PhD students in-person, after joining the program 5 months ago. Thankfully, three of them had already visited me in Athens, which made the initial apprehension of facing a group of 15 new (but amazing) people much easier.  The first two days of talks proved intense and demanded all the brainpower we could muster. But once the talks were over, we still had the perfect opportunity to replenish it, just as Ilpo Vattulainen advised, by eating Alsacian-style fish during the planned social events. Nonetheless, it was interesting to hear firsthand about the current role of allostery in research, both from the academic and pharmaceutical perspectives, after having spent countless hours reading about it in my first months of PhD. On the third day, I was finally able to hear my colleagues explain their projects and share their progress in their own words. It is always alluring to learn about science outside one's own tiny research niche, and to witness the seamless flow of discussions that arose between different fields of research. Attending the ESR-exclusive training activities (and equally importantly, our own unplanned socialization events after) was an invaluable opportunity to share and discuss between us about the challenges and achievements of undertaking a PhD, and find support in one another. I appreciated the chance to get to know my peers on a more personal level, exchanging stories and experiences, and realizing how serendipity brought us all together from different places and backgrounds. As my TGV departed from Strasbourg, I was left with a pleasant sense of fulfillment and growth. And an even more intense urge to meet everyone again in just 4 months! See you soon. Using EPR spectroscopy in the study of protein conformations - a blogpost by ESR6 Sigrid Pedersen10/4/2023 One of the main issues with obtaining protein structures, especially that of transmembrane proteins, are their relative flexibility. This issue limits both of the most commonly used methods of obtaining larger protein structures: X-ray crystallography and cryo-electron microscopy (cryo-EM). Indeed, in their native environment, proteins are highly dynamic and cycle through different conformations depending on their immediate surroundings. However, X-ray crystallography can only obtain one protein conformation at a time and even the number of conformation obtained by cryo-EM is limited. Intermediary structures between the active and inactive form of a protein are particularly difficult to sample, resulting in a considerable bottleneck in the analysis of conformational changes.
Luckily, one particular spectroscopy method, pioneered 25 years ago by Altenbach et al. and which requires the insertion of paramagnetic probes, can help feel in the gaps. This Electron Paramagnetic Resonance (EPR) utilizes the ability of unpaired electrons to change between their two spin states (ms = ½ or ms = - ½) and the influence that the nearby environment and associated atoms has on them to derive a spectral line. One particularly useful application of this method for protein structural analysis is the double electron-electron resonance (DEER) measurements: the distance between a pair of labelled spins is measured, and then that distance can be measured again for the same protein under different conditions (for example in the presence of various ligands). Nowadays, the most common way to establish spin labeling is by inserting reactive cysteines via site-selective mutagenesis at dynamically relevant sites in the protein of interest. The unpaired electrons within the accessible cysteines will then bind to the spin labels when incubated with the purified proteins under specific conditions. My own project at the Scheerer laboratory will include performing this experiment on the melanocortin-4 receptor (MC4R) in different environments, such as in the presence of orthosteric and allosteric ligands, in collaboration with Matthias Elgeti’s laboratory at Leipzig University. So far, I am pleased to report that the wildtype MC4R has been successfully labeled by both MTSL (S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate) and IDSL (bis(2,2,5,5-tetramethyl-3-imidazoline-1-oxyl-4-il)-disulfide spin label). MTSL is a nitroxide reagent with a high reactivity towards cysteines. Its high labeling efficiency combined with its small size, which minimizes the risk of interference with the protein structure and the folding pathway, have made it the most popular spin label for proteins. The reactive disulfide IDSL, on the other hand, labels cysteines via sulfhydryl-disulfide exchange reaction. Additionally, an intra-side chain S-N stabilizes the interaction, making IDSL an attractive label for DEER measurements. The next step is now to obtain a “cysteine-less” variant, which cannot be labeled for EPR, as a negative control. This represents one of the unfortunate drawbacks of EPR spectroscopy, as this process is time-consuming, and we have to account for the unpredictable possibility that this might result in a loss of expression or structure. This is what I am currently doing, generating two virus containing “cysteine-less” mutants of MC4R: one of them had four reactive cysteines mutated into serines and the other had five (the same four and one additional cysteine that is believed to undergo post-translational modification in humans). My hope is that at least one of these MC4R mutants will not only be expressed in a large enough amount to be used in subsequent experiments, but also that EPR will show no labelling of the remaining cysteines. Indeed, we consider that the remaining native cysteines, being either involved in a disulfide bond or hidden within the transmembrane core, should not be available for labeling. If this experiment is successful, I will then be able to move on to constructs with cysteines inserted at interesting sites, such as in the transmembrane regions 5, 6 and 7 and on the intracellular loop 2, and thus begin the DEER measurements in earnest. Time goes by so fast, that’s undeniable. I remember the day I started my PhD as if it was yesterday. Here I am today, almost in the second year of PhD. At this pace, I have no doubt that in the blink of an eye, I will be writing my thesis.
When I look back, what I can say is this last year was extremely unique and memorable with its highs and lows. Well, you more or less know about my journey from my previous blogpost, but what I hadn’t told you, also what I didn’t know, at that moment was the challenges that a computational chemist has to go through topped with the mysterious world of experiments, not only in academia but also in the corporate world. Now please bear with me and let me tell you the tale of the “shapeshifter” (that’s what I like to call myself lately) As someone who chose the computational chemistry path back in the sophomore year of college, I was slowly straying away from the idea of performing experiments in the lab again. My practical lab courses were long gone. As the time passed by, my skills regarding experimental methods got rusty more and more. After I had an interview to get the ESR4 position, Prof. Carles Curutchet asked me whether or not I would also be willing to do some experiments for the project. I thought it would be great to do both the experimental and the computational part of the job to be able to own it completely. Though I cannot deny that I was a little scared. As I was gaining more experience and deepening my knowledge in theoretical methods, my ability in performing experiments in the lab was decaying at the same pace. I was afraid of getting lost in things which could be pretty obvious to some wet chemists but clearly not to me, terrified by even the thought of failing… As human beings, when it comes to unknown things, we are always a bit anxious and scared, aren’t we? It is because we don’t exactly know what we are facing and how to deal with it, but it is also somewhat intriguing and exciting, don’t you agree? So, long story short, I didn’t chicken out and said yes to performing experiments besides the computational work I was supposed to do. In a few months, I found myself re-learning the fundamentals of working in a lab (general chemistry lab 101 reloading…) I was cautious as if I could possibly press a button and burn the whole lab down. I asked for guidance, talked to the PhD students, professors, read the manuals of the machines I was going to use before taking action. For each type of experiment, I was supposed to use a specific machine which was located in different labs of the department. So, in the morning I was in one lab, in the afternoon in another one. Even people seeing me here and there were not entirely sure which lab group I belonged to or who I was working with. After some time, in the experimental section of the department, I became someone everyone was used to seeing around but nobody knew much about, and little did they know that I was actually a member of the computational chemistry group. Having two different roles also made it complicated to explain what exactly I was doing. Yet, it was just the tip of the iceberg. In February, I started my secondment at Gain Therapeutics. The company was located only 5 mins away from the pharmacy campus of University of Barcelona where I normally work at. So, in a way it was different than the usual secondment concept where you move to another country for a couple of months and work at another institution while a new culture is being introduced to your system. For me it was regular PhD work + secondment in the same city all blended in together. There were days I stopped by the university in the morning to get a sample and then went to work. It just added one more location to the whole equation of where I work. In the company, I got a nice desk in a nice office, like any other member of the computational team, but as a part of my secondment plan, I was also supposed to perform some experiments in the Gain lab which was completely on the opposite side of the office. After going down with the elevator, taking a long walk down the hallway, passing through two turnstiles, scanning your card twice, opening several doors, congratulations you made it to the lab (In this case is it short for laboratory or labyrinth). After a short while, I figured out that the machine they had in the Gain lab was not suitable for my experiments. So, I had to use the one in the common area which was one story below the Gain lab and required going through another door with scanning your card, though that door required another type of authorization which I didn’t have as a visiting researcher. So, every time I had to use a machine, I either needed someone to accompany me or borrow someone else’s card. While working there in the lab, I got into the world of biologists, because most people working in the lab had a biology background. Actually, they were surprised when I told them I was a chemist (just a chemist, let alone the computational part). Here I am now, performing some experiments + simulations at academia & company. I can be a computational or an experimental chemist or both, I can be at the university or appear at the company or both. It feels like I have some magical powers and I can shapeshift from one to another like a druid (gamer detected). So, at this point I am not sure how to explain where I work, what I do exactly, which team I belong to. When I get asked such questions, oh I can guarantee that there is a long story on its way, but to keep it shorter and make it sound more like me I will summarize my current PhD life with an acrostic: Little did I know that I was about to start one hell of a journey I thought I’d just be on my PC without getting my hands dirty Fast-forward a few months, I started to conduct an experiment Eventually I found myself at Gain exploring company environment One day you can find me around the spectrometer in the lab For the rest, I am doing my simulations at the office looking fab Partially a student at school, partially a researcher at the company Half wet chemist, half computational, not wearing just one hat, but so many Don’t underestimate the life of a comperimental chemist in academpany |
- Home
- People
- Research
-
Training
-
Training Events
>
- 1st Workshop and PhD Induction Course
- 1st Training School & Networking Meeting
- Allostery in Drug Discovery Awareness Event and Symposium
- 2nd Training School & Networking Meeting
- IPR Training for Researchers & ESR Presentations on the Progress of their Research
- 3rd Training School & Networking Meeting
- European Conference in Allostery in Drug Discovery
- Secondments
- ALLODD Webinars
- Courses & Lectures
- Journal Club
-
Training Events
>
- Dissemination
- Events
- Newsroom
- Job Openings
- Blog
- Contact




 RSS Feed
RSS Feed
